|
 |
| J Korean Acad Pediatr Dent > Volume 48(3); 2021 > Article |
|
ņ┤łļĪØ
ņØ┤ ņŚ░ĻĄ¼ņØś ļ¬®ņĀüņØĆ ļ░£Ļ▒░ ļÉ£ ļ¦żļ│Ą ņāüņĢģ Ļ│╝ņ×ēņ╣śņŚÉņä£ ņ¢╗ņØĆ ņ╣śņłśņ£Āļל ņżäĻĖ░ņäĖĒżņØś ņ┤łĻĖ░ Ļ│äļīĆņÖĆ ĒøäĻĖ░ Ļ│äļīĆņØś ņāüņĢäņ¦łļ¬©ņäĖĒż ņ£ĀņĀäņ×ÉņØś ĒŖ╣ņä▒ ņØä ņĢīņĢäļ│┤ļŖö Ļ▓āņØ┤ļŗż.
ņĀäņŗĀ ņØśĻ│╝ ļ│æļĀźņØ┤ ņŚåļŖö 6 - 9ņäĖ ņé¼ņØ┤ņØś ļé©ļģĆņĢäņØ┤ 12ļ¬ģņŚÉĻ▓īņä£ ņä£ļ®┤ļÅÖņØśļź╝ ņ¢╗Ļ│Ā ļ¬©ļæÉ ņāüņĢģņŚÉ ņ£äņ╣śĒĢ£ Ļ│╝ņ×ēņ╣śļź╝ ļ░£Ļ▒░ĒĢśņŚ¼ ļŗ╣ņØ╝ ļ░£Ļ▒░ ļÉ£ Ļ│╝ņ×ēņ╣śņØś ņ╣śņłśņäĖĒżļź╝ ņ▒äņĘ©ĒĢśņśĆļŗż. 12Ļ░£ņØś ņäĖĒżļź╝ Ļ░üĻ░ü 3Ļ│äļīĆņÖĆ 10Ļ│äļīĆņŚÉņä£ Ļ│©ĒśĢņä▒ ņ£ĀļÅä ļČäĒÖöņĀ£ļź╝ ņ▓śļ”¼ĒĢ£ ĻĄ░Ļ│╝ ņ▓śļ”¼ĒĢśņ¦Ć ņĢŖņØĆ ĻĄ░ ņØä ļéśļłäņ¢┤ ņŗżņŗ£Ļ░ä ņżæĒĢ®ĒÜ©ņåī ņŚ░ņćäļ░śņØæņØä ņŗ£Ē¢ēĒĢśņŚ¼ ņāüņĢäņ¦łļ¬©ņäĖĒżņØś ĒŖ╣ņä▒ņØä ņĢīņĢäļ│┤ņĢśļŗż. ņé¼ņÜ®ļÉ£ ņ£ĀņĀäņ×ÉļŖö osteonectin (ONT), alkaline phosphatase (ALP), osteocalcin (OCN), dentin matrix protein 1 (DMP-1), ĻĘĖļ”¼Ļ│Ā dentin sialophosphoprotein (DSPP)ņśĆļŗż.
ņ£ĀņĀäņ×É ļ░£Ēśäņ¢æņØĆ, ļČäĒÖöņĀ£ļź╝ ņ▓śļ”¼ĒĢśņ¦Ć ņĢŖņØĆ ĻĄ░ 3Ļ│äļīĆņŚÉņä£ļŖö ONT, ALP, OCN, DMP-1, DSPPņł£ņä£ļĪ£ ļ¦ÄņØ┤ ļ░£ĒśäĒĢśņśĆļŗż. ļČäĒÖöņĀ£ļź╝ ņ▓śļ”¼ĒĢś ņ¦Ć ņĢŖņØĆ ĻĄ░ 10Ļ│äļīĆņŚÉņä£ļŖö ONT, DMP-1, OCN, ALP, DSPPņł£ņ£╝ļĪ£ ONT, OCN, DSPPņØś ņł£ņä£ņŚÉļŖö ļ│ĆĒÖöĻ░Ć ņŚåņ¦Ćļ¦ī ALP, DMP-1ņØś ņł£ņä£ļŖö ņä£ļĪ£ ļ░öļĆīņŚłļŗż.
ņØ┤ņāüņØś Ļ▓░Ļ│╝ļź╝ ņóģĒĢ®ĒĢ┤ ļ│╝ ļĢī, ALPņÖĆ DMP-1ņØĆ 3Ļ│äļīĆņÖĆ 10Ļ│äļīĆ ņäĖĒżņØś ļČäĒÖöļź╝ ņ£äĒĢ£ ņżæņÜöĒĢ£ Ēæ£ņ¦Ćņ×ÉļĪ£ ņé¼ņÜ®ļÉĀ ņłś ņ׳ļŗż. Ļ│╝ņ×ēņ╣ś ņ╣śņłśņ£Ā ļל ņżäĻĖ░ņäĖĒżļŖö ņāüņĢäņ¦łļ¬©ņäĖĒżņØś ĒŖ╣ņä▒ņØä Ļ░Ćņ¦Ćļ®░, ļśÉĒĢ£ Ļ│╝ņ×ēņ╣śĻ░Ć ņ¢┤ļ”░ ļéśņØ┤ņŚÉ ļ░£Ļ▒░ļÉśĻ│Ā 10Ļ│äļīĆĻ╣īņ¦Ć ņåīņÜöļÉśļŖö ņŗ£Ļ░äņØ┤ ņĀüĻ▓ī Ļ▒Ėļ”░ļŗżļŖö Ļ▓āņØä Ļ│ĀļĀżĒĢśļ®┤, Ļ│╝ņ×ēņ╣śļŖö ņ╣śņĢä ņ£Āļל ņżäĻĖ░ņäĖĒżņØś Ļ│ĄņŚ¼ļČĆļĪ£ņä£ ĒøīļźŁĒĢ£ ĒÖ£ņÜ®Ļ░ĆļŖźņä▒ņØ┤ ņ׳ņØīņØä ĒÖĢņØĖĒĢśņśĆļŗż.
Abstract
The aim of this study is to compare the properties of odontoblast gene of early passage cells and late passage cells derived from impacted maxillary supernumerary teeth.
Impacted supernumerary teeth with maxilla were extracted from 12 patients (8 males, 4 females) between 6 - 9 years old without medical history. Real-time polymerase chain reaction (PCR) was conducted to compare characterization of odontoblast cell in the 3rd and 10th passage, and between with bone inducing additive group and without additive group. Genes for odontoblasts characteristics are osteonectin (ONT), alkaline phosphatase (ALP), osteocalcin (OCN), dentin matrix protein 1 (DMP-1) and dentin sialophosphoprotein (DSPP).
The level of gene expression was in a decreasing order of ONT, ALP, OCN, DMP-1 and DSPP in the 3rd passage, and in decreasing order of ONT, DMP-1, OCN, ALP, and DSPP in the 10th passage in the undifferentiation and differentiation group. The order of ONT, DMP-1, and OCN did not changed. ALP and DMP-1 were switched in order.
ALP and DMP-1 may be used as important markers for differentiating between the 3rd passage and 10th passage cells.
Considering that supernumerary tooth was extracted young age and the time required to cultured 10th passage was short, supernumerary tooth can be considered a useful donor site of dental pulp stem cells.
Dentin has a pulp protection feature to form secondary dentin. The organic contents of dentin are mainly type 1 collagen and noncollagenous matrix protein[1].
During the differentiation from ectomesenchymal cell to preodontoblast to odontoblast, specific genes such as osteonectin (ONT), alkaline phosphatase (ALP), osteocalcin (OCN), dentin matrix protein 1 (DMP-1) and dentin sialophosphoprotein (DSPP) are secreted[2-4].
Various studies have been conducted on the source of stem cells related to teeth. Representative ones are wisdom teeth[5,6], deciduous teeth during the shedding period[7], and supernumerary tooth[8]. Wisdom teeth were usually extracted during adolescence, and passaging of dental pulp stem cells derived from wisdom teeth usually takes 7 - 10 days[9]. In comparison, passaging of dental pulp cells derived from supernumerary teeth takes 3 - 4 days[10]. Supernumerary teeth are usually present in the midline between the two maxillary central incisors, and are more abundant than natural teeth[11]. It is recommended to be extracted at early age as they may disrupt the eruption of adjacent teeth and induce the formation of dentigerous cyst[12,13]. Therefore, research on supernumerary teeth as a source of tooth-derived dentin pulp stem cells is necessary.
Pulp derived stem cells were able to proliferate up to 20 - 30 population doublings (PD) and maintain immunosuppressive ability[14]. More cells can be obtained from limited sources through subculture. One study reported no difference in the differentiation of mesenchymal stem cells to osteoblasts in different passages[15], whereas another study reported reduced multilineage developmental potential of marrow stromal cells as they were passaged at high concentrations[16]. However, only few studies have compared the characteristics of early and late passage cells.
This study aimed to compare the characteristics of early passage cells (3rd passage) and late passage cells (10th passage) derived from supernumerary tooth using real-time polymerase chain reaction (PCR), and investigate potential usefulness of these characteristics.
12 patients aged 6 - 9 years without a medical history of systemic disorders, who visited the clinic for extraction of supernumerary teeth, participated in this study (Table 1). Extraction of supernumerary teeth for the purpose of collection of dentin pulp cells was approved by the Institutional Review Board of Dankook University Dental Hospital (H-1506/006/001), and was performed under the consent of the patients and their legal guardians.
The human dental pulp cells from 12 patients (8 males, 4 females) were run on real time-PCR. The patients underwent radiographic evaluation and/or computed tomography. Extraction was performed by an experienced technician.
Collected tissues were stored in an ╬▒-minimum essential medium (╬▒-MEM, Gibco BRL, Grand Island, NY, USA) containing 100 U/mL of penicillin, 100 ╬╝g/mL streptomycin (Gibco BRL, Grand Island, NY, USA), 2 mM L-glutamine (Gibco BRL, Grand Island, NY, USA), 10 mM L-ascorbic acid (Sigma, St. Louis, MO, USA) and fetal bovine serum (FBS, Gibco, Life Technologies Corporation, Carlsbad, Calif., USA) immediately after the tooth extraction to minimize cell loss. They were then moved to the laboratory for collection of human dental pulp cells.
For cell extraction, a sterile high-speed cutting machine was used to section the supernumerary teeth below the cementoenamel junction under sterile distilled water without exposing the dental pulps. Teeth were then broken to expose the dental pulps. The exposed dental pulps were collected using a sterile file.
Supernumerary dental pulp stem cells (sDPSCs) were obtained via enzymatic digestion of the collected pulp tissues[17]. After cutting the tissues into 1 mm3 pieces, they were placed in a shaking incubator at 37┬░C with 3 mg/mL of type 1 collagenase (Sigma-Aldrich Co., St. Louis, M.O., USA) and 4 mg/mL of dispase (Sigma-Aldrich Co., St. Louis, M.O., USA) for 1 hour for cell separation. Next, cells isolated with a 70 ╬╝m Falcon strainer (CORNING Inc., N.Y., USA) were filtered and cultured in a ╬▒-MEM medium containing 20% FBS (Gibco, Life Technologies Corporation, Carlsbad, Calif., USA), 100 U/mL penicillin, 100 ╬╝g/mL streptomycin (Gibco BRL), 2 mM L-glutamine (Gibco BRL) and 10 mM L-ascorbic acid (Sigma, St. Louis, MO, USA) at 37┬░C and 5% CO2 for 4 - 6 days until 70 - 80% confluency was achieved. The obtained cells were named sDPSCs. The medium was replaced every 2 - 3 days starting 48 hours after the primary culture. Any floating materials that did not attach to the growth medium were rinsed off with phosphate buffer saline. When the 80% of the medium was covered by cells, the cells were isolated using trypsin-EDTA (CORNING Inc., N.Y., USA). Then, the medium was divided into four sections, in each of which the cells were subcultured for 10 passage.
To evaluate the differentiation of supernumerary tooth-derived dental pulp stem cells into osteoblastoma, osteoblastoma were cultured in growth media until the 3rd passage or 10th passage. To induce differentiation into osteoblastoma, each sample of stem cells was cultured in a growth medium containing a mixture of 10mL ╬▒-MEM and 10% FBS (Gibco, Life Technologies Corporation, Carlsbad, Calif., USA), and a mixture of 5 mM ╬▓-glycerophosphate (Sigma-Aldrich Co., St. Louis, M.O., USA), 100 nM dexamethasone (Sigma-Aldrich Co., St. Louis, M.O., USA) and 100 ╬╝M ascorbic acid (Sigma-Aldrich Co., St. Louis, M.O., USA). Each medium was replaced every 3 days, and differentiation was induced for 8 days[10].
Total RNA extraction was performed using Easy-spin total RNA extraction Kit (iNtRON Biotechnology, Gyeonggi-do, Korea). Then, Nanodrop ND-2000┬« (Thermo Scientific, Waltham, MA, USA) spectrophotometer was used to quantify total RNA. For synthesis of cDNA, 1 ┬Ąg of total RNA was added to quantitative polymerase chain reaction reverse transcription (qPCR RT). Master Mix kit (TOYOBO Co., Osaka, Japan) containing oligo (dT) 15 primer, and nuclear-free water was added to make the total volume 20 ┬Ąl. Then, PCR was ran for 5 minutes at 65┬░C, 5 minutes at 4┬░C, and 5 minutes at 95┬░C.
To study patterns of mRNA expression through real-time PCR, 4 samples were prepared: additive-treated in 3rd passage and 10th passage cells (differentiation group), and non-additive-treated in 3rd passage and 10th passage cells (undifferentiation group). Then, mRNA expression patterns of the ALP, OCN, ONT, DMP-1, and DSPP genes, which are associated with hard tissue formation, were analyzed. The comparison of levels of mRNA expression before and after the cell differentiation was made relative to the level of glyceraldehyde 3-phosphate dehydrogenase (GADPH) mRNA expression. cDNA was diluted with sterile distilled water until the total mass of the sample reached 25 ng. Then, the sample was reacted with 0.5 ╬╝L of forward and reverse primers (10 pmol/╬╝L) each and 2 ├Ś 5 ╬╝L SYBR Premix Ex TaqŌäó (Bio-Rad Laboratories, Hercules, C.A., USA) in StepOnePlusŌäó Real-time PCR (AB Applied Biosystems by life Technology, Waltham, M.A., USA) for amplification and fluorescence detection. The cDNAs were denatured at 95┬░C for 20 seconds, and annealed at the optimal temperature of the primers for 1 minute. This process was repeated for 40 cycles. Next, the cDNAs were dissociated at 65 - 95┬░C, and their melting curve was analyzed. Whether the melting curve exhibited a single maximal point was checked, indicating production of only one PCR product. The conditions of the primers used in the study are shown in Table 2. Collected data were analyzed using StepOnePlusŌäó (AB Applied Biosystems by life Technology, Waltham, M.A., USA).
The real-time PCR samples were statistically analyzed 3 times each. The mean and standard errors were calculated for each sample. For comparison of relative levels of mRNA of each gene, a 2-Ō¢│Ō¢│CT method was used with GAPDH as the internal control gene[18]. To compare relative levels of mRNA between the 3rd passage and 10th passage in the differentiation and undifferentiation groups, a Mann-Whitney U test (p < 0.05) was performed using SPSS (version 23.0, Chicago, USA). Differences in the relative levels of mRNA of each gene before and after the additive treatment in the 3rd passage and 10th passage were found by performing a Wilcoxon signed rank test (p < 0.05) using SPSS (version 23.0, Chicago, USA).
The media were 70 - 80% confluent with sDPSCs after 4 - 6 days of primary culture, and the sDPSCs were subcultured to the next passage.
Ct values of the ALP, OCN, ONT, DMP1, and DSPP genes, as well as GAPDH used as the housekeeping gene were obtained. Then, Ō¢│Ct (threshold cycle) values (target gene - housekeeping gene) were calculated to find the relative levels of gene expression. Fig. 1 shows the Ō¢│Ct values for 3rd and 10th passage group in the undifferentiation group. The smaller the Ō¢│Ct value, the higher the number of genes included in the specimen. In 3rd passage, Ō¢│Ct value of the ONT gene was the lowest, and that of the DSPP gene was the highest in both the 3rd and 10th passage. In the undifferentiation group, Ō¢│Ct values of the OCN, ONT, and DMP-1 genes decreased, and those of the ALP and DSPP genes increased in both of 3rd and 10th passage.
Mann-Whitney U test showed no significant difference in the level of expression of the ALP gene, and significant differences in the levels of all the other proteins between the two passages (p < 0.05). Among the four genes that showed significant differences between the two passages, only the DSPP gene was expressed in a decreasing number of cells as they were subcultured until the 10th passage, while the OCN, ONT, and DMP-1 genes were expressed in a greater number of cells.
Fig. 2 shows the Ō¢│Ct values for 3rd and 10th passage group in the differentiation group. In the differentiation group, Ō¢│Ct value of the ONT gene was the lowest, and that of the DSPP gene was the highest in both the 3rd and 10th passage. Ō¢│Ct values decreased for the OCN, ONT, DMP-1, and DSPP genes, while that of ALP increased slightly. Mann-Whitney U test showed no significant differences in the level of expression of the ALP gene between the two passages (p < 0.05), whereas significant differences in that of the OCN, ONT, DNP-1 and DSPP genes were found between the two passages.
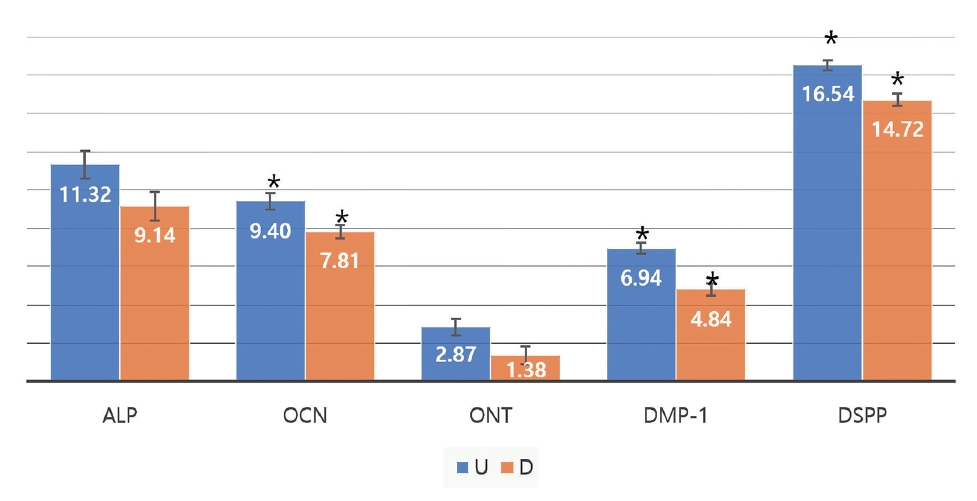
Fig. 3 shows the Ō¢│Ct values before and after additive treatment for 3rd passage group. As a result of comparing before and after additive treatment at 3rd passage, Ō¢│Ct values decreased for the ALP, OCN, ONT, and DMP-1 genes, while that of DSPP increased.
Wilcoxon signed rank test showed significant differences in the level of expression of the ALP, DMP-1, and DSPP genes before and after the additive treatment in the 3rd passage (p < 0.05). No significant difference was found in the level of expression of the OCN and ONT genes (p > 0.05). Therefore, the level of expression of the ALP and DMP-1 genes increased, and that of the DSPP gene decreased after the additive treatment in the 3rd passage.
Fig. 4 shows the Ō¢│Ct values before and after additive treatment for 10th passage group. At 10th passage, every Ō¢│Ct values decreased after additive treatment.
Wilcoxon signed rank test showed statistically significant differences in the levels of expression of the OCN, ONT, DMP-1, and DSPP genes before and after the additive treatment (p < 0.05). No significant difference in the level of expression of the ALP gene was found (p > 0.05). Therefore, the level of expression increased for all genes except for the ALP gene in the 10th passage.
Table 3 shows the order of relative levels of gene expression. The level of gene expression was in a decreasing order of ONT, ALP, OCN, DMP-1 and DSPP in the 3rd passage, and in decreasing order of ONT, DMP-1, OCN, ALP, and DSPP in the 10th passage in the undifferentiation group. While the order of ONT, OCN and DSPP did not changed, ALP and DMP-1 were switched in order. Similar observation was made in the differentiation group.
Comparing the results before and after differentiation, in 3rd passage, gene expression was in a decreasing order of ONT, ALP, OCN, DMP-1, DSPP in both differentiation, undifferentiation group. Also, in 10th passage, gene expression was in a decreasing order of ONT, DMP-1, OCN, ALP, DSPP in both differentiation, undifferentiation group. Meanwhile, in total, ONT, OCN, DSPP had no change in order, but the order of ALP and DMP-1 changed.
The purpose of this study is to find out the characteristics between relatively early passage (3rd passage) and late passage (10th passage) using real-time PCR in supernumerary dental pulp stem cells derived from maxillary supernumerary tooth.
Since Gronthos et al .[5] revealed that adult stem cells obtained from the pulps of erupted wisdom teeth have characteristics of bone marrow derived stem cells, teeth have become a research target of many studies as a source of adult stem cells. Examples of stem cells may be obtained include stem cells from exfoliated deciduous teeth (SHED)[7], periodontal ligament stem cells (PDLSCs)[19], dental follicle precursor cells (DFPCs)[20], stem cells from apical papilla (SCAP)[21], and supernumerary teeth[8].
The prevalence rate of supernumerary teeth varies from 1.0 - 3.5%. Supernumerary teeth must be extracted at early age as they may cause pathological problems due to delayed eruption of adjacent permanent teeth, orthodontic problems related to teeth rotation, and formation of dentigerous cysts[22]. Extraction of supernumerary teeth provides an opportunity to obtain young stem cells from these teeth.
Therefore, in this study, cells of 3rd passage, which are most widely used in adult stem cell studies, and cells of 10th passage, which are considered to be relatively late, were obtained and treated with additive for 8 days to induce hard tissue formation. To confirm the characteristics of each of passage, and before and after differentiation, the amount of mRNA gene expression of ALP, OCN, ONT, DMP-1 and DSPP mainly found in odontoblasts was measured using real-time polymerase chain reaction analysis.
The value obtained from the real-time polymerase chain reaction is the threshold cycle. This value refers to the number of cycles in the fluorescence signal representing the threshold value and the contact point due to the significant increase in the amplification curve[18]. In this study, the results were analyzed using the relative quantification method proposed by Livak and Schmittgen[18]. GAPDH was used as a housekeeping gene.
In study comparing stem cells derived from pulp cells and periodontal ligament cells, the pulp cells expressed DSPP, whereas the periodontal ligament cells did not express DSPP[23].
In addition, after Gronthos et al .[14] found that pulp-derived stem cells can proliferate over at least 20 - 30 population doublings, can safely be stored frozen, and have immunosuppressive properties, more cells can be obtained through subculture and the range of utilization has increased. However, there was also a previous study revealing that when the mesenchymal stem cells were subcultured at a high concentration, the cell morphology was same as the passage increased, but the pluripotency of the cells decreased[16]. In case of mesenchymal stem cells, after treatment with a differentiating agent that induces hard tissue formation, there was little effect from subculture, but there were conflicting opinions that the differentiation ability decreased by subculture in inducing differentiation into adipocytes[24].
Dentin can form secondary dentin even after differentiation is over. Odontoblast plays the main role[25]. During the differentiation from ectomesenchymal cell to preodontoblast to odontoblast, preodontoblast mainly form ONT, ALP, and OCN[26], and differentiated odontoblast form DSPP, DMP-1, DMP-2, DMP-4, bone sialoprotein (DSP), matrix extracellular phosphorylated protein (MEPE) and osteopontin (OPN)[2,3].
ONT is a phosphorylated glycoprotein synthesized by various cells including osteoblasts[27]. ONT is observed in non-calcified tissues such as the periodontal ligaments and pulps of pig and cow[28]. In human, the ONT gene is expressed during early stages of differentiation into osteoblasts, or in preosteoblasts[4]. Therefore, it is expressed to a greater degree in preodontoblasts within he tooth germ than in erupted teeth[29]. In this study, the ONT gene was expressed the most in both the 3rd and 10th passage. This is consistent with the findings of previous studies in which the ONT gene was expressed at relatively early stages of differentiation. The level of expression of the ONT gene increased 8.03 folds in the undifferentiation group and 11.40 folds in the differentiation group from the 3rd passage to 10th passage. The level of expression of the ONT gene increased 1.94 folds in the 3rd passage, and 2.82 folds in the 10th passage after the additive treatment. Since the level of expression of the ONT gene did not significantly increase when differentiation into odontoblasts was induced, the result of this study is consistent with those of previous studies, which reported that ONT is synthesized in preodontoblasts.
ALP is an ectoenzyme consisting of glycoproteins, which are present in various tissues within the body[30]. Its expression increases during the formation of mineralized tissues[31]. ALP gene was decreased to 0.71 times and 0.84 times the level in the 3rd passage in the undifferentiation and in the differentiation group, respectively. The level of expression of the ALP gene increased 8.91 folds and 4.52 folds in the 3rd passage and 10th passage, respectively, after the additive treatment. As can be seen, the level of expression of the ALP gene was not significantly affected as the number of passage increased, and significantly increased after additive treatment.
OCN is a biomarker found after cementoblast and odontoblast differentiation, and can be used to evaluate the degree of differentiation. It also controls the degree of mineralization[32,33]. In this study, the OCN gene was expressed to the 3rd highest degree among the five genes of interest in both the 3rd and 10th passage. This supports the results of previous studies that the OCN gene is expressed after cells have differentiated into odontoblasts, and that OCN may be used as a marker for measuring the degree of differentiation. The level of expression of the OCN gene increased 7.42 folds and 14.29 folds between the 3rd passage and the 10th passage in the differentiation and undifferentiation group, respectively. Therefore, the level of expression of the OCN gene increased as the cells reached the 10th passage. The level of expression of the OCN gene increased 1.56 folds and 3.01 folds in the 3rd passage and 10th passage, respectively, after the additive treatment. The level of OCN gene expression was not significantly affected by the additive treatment.
DMP-1 is an acid phosphatase found in the dental pulp. It is not found in osteoblasts, and is characteristically produced in bone cells embedded in the bone matrix[34]. It usually appears after the bud stage unlike DSPP, which appears at the cap stage[35]. In this study, the level of DMP-1 gene expression increased 323.54 folds between the 3rd passage and the 10th passage in the undifferentiation group, marking the highest increase among the five genes of interest. It also increased 494.14 folds in the differentiation group. DMP-1 may be used as an important marker during identification of characteristics of cells through different stages of passaging in future studies. The level of expression of the DMP-1 gene increased 2.81 folds and 4.30 folds in the 3rd passage and 10th passage, respectively, after the additive treatment. Therefore, the level of DMP-1 gene expression was not significantly affected by the additive treatment.
DSPP is a non-collagenous protein, and a prominent example of pulp-specific gene[36]. Dentin sialoprotein (DSP), dentin phosphoprotein (DPP) and dentin glycoprotein (DGP) are all synthesized from the DSPP gene. DPP is a product of C-terminal specific DSPP degradation, and DSP is a product of N-terminal specific DSPP degradation[37]. DSPP induces calcification of the dental pulp along with ALP and OCN[36,37]. In this study, the level of expression of the DSPP gene was reduced to 0.39 times the level in the 3rd passage in the 10th passage in the undifferentiation group. In the differentiation group, the level increased 2.72 folds in the 10th passage. Since the absolute level of expression of the DSPP gene was the lowest among the five genes of interest, it can be predicted that its level of expression will increase after the 10th passage. In the 3rd passage, the level of expression of the DSPP gene was reduced to 0.50 times the initial level after the additive treatment. The level increased 3.55 folds in the 10th passage after the additive treatment. It was therefore observed that the level of DSPP gene expression tended to decrease in the 3rd passage, and then increase in the 10th passage.
The level of mRNA expression was in decreasing order of ONT, ALP, OCN, DMP-1, and DSPP in the 3rd passage, and ONT, DMP-1, OCN, ALP, and DSPP in the 10th passage. While the order of ONT, OCN, and DSPP was unchanged, ALP and DMP-1 were switched to the 4th and 2nd highest, respectively. A similar pattern was observed for the differentiation group. Regardless of the treatment with differentiation agents, ONT, OCN, and DSPP were the 1st, 3rd, and 5th most expressed, and the order of ALP and DMP-1 was changed in the 2nd and 4th order. From this, it is thought that ALP and DMP-1 could be important markers that distinguish between the 3rd and 10th passage.
Based on the findings above, sDPSCs have characteristics of odontoblasts in both the 3rd and 10th passage. Therefore, sDPSCs may be useful in studies in which differentiation into a certain type of tissue is induced by making use of the passage-specific expression of proteins.
This study investigated the degree to which tooth-derived dental pulp stem cells exhibit characteristic gene expression of odontoblasts before and after additive treatment in 3rd and 10th passage using real-time PCR. Gene expression of these stem cells is conserved across both passages, and that the two passages can be differentiated based on the level of expression of the ALP and DMP-1 gene.
Based on this finding that sDPSCs have gene expression patterns of odontoblasts in both the 3rd and 10th passage, wide application of sDPSCs may become possible after sufficient research conducting their potential to differentiate into other types of tissues other than teeth.
Fig┬Ā1.
Mean value of Ō¢│Ct (X-H) between 3P and 10P in undifferentiation.
p value from Mann-Whitney U test
ONT: Osteonectin, ALP: Alkaline phosphatase, OCN: Osteocalcin, DMP-1: Dentin Matrix Protein 1, DSPP: Dentin sialophosphoprotein, P: Passage, Ō¢│Ct(X-H): Target gene - Housekeeping gene

Fig┬Ā2.
Mean value of Ō¢│Ct (X-H) between 3P and 10P in differentiation.
p value from Mann-Whitney U test
ONT: Osteonectin, ALP: Alkaline phosphatase, OCN: Osteocalcin, DMP-1: Dentin Matrix Protein 1, DSPP: Dentin sialophosphoprotein, P: Passage, Ō¢│Ct(X-H): Target gene - Housekeeping gene
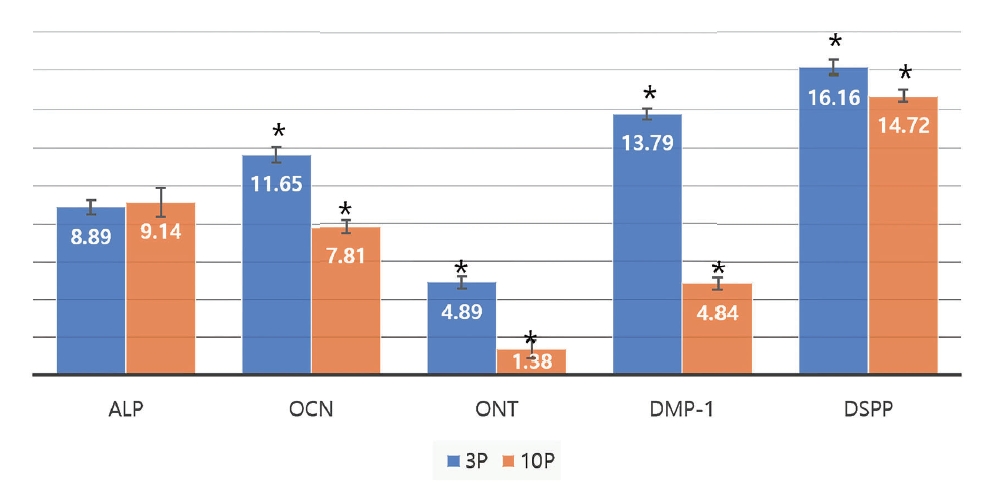
Fig┬Ā3.
Mean value of Ō¢│Ct (X-H) between differentiation and undifferentiation in 3rd passage.
p value from Wilcoxon signed rank test
ONT: Osteonectin, ALP: Alkaline phosphatase, OCN: Osteocalcin, DMP-1: Dentin Matrix Protein 1, DSPP: Dentin sialophosphoprotein, P: Passage, Ō¢│Ct(X-H): Target gene - Housekeeping gene

Fig┬Ā4.
Mean value of Ō¢│Ct (X-H) between differentiation and undifferentiation in 10th passage.
p value from Wilcoxon signed rank test
ONT: Osteonectin, ALP: Alkaline phosphatase, OCN: Osteocalcin, DMP-1: Dentin Matrix Protein 1, DSPP: Dentin sialophosphoprotein, P: Passage, Ō¢│Ct(X-H): Target gene - Housekeeping gene
Table┬Ā1.
Baseline characteristics of patient population
Table┬Ā2.
Annealing temperatures for the primer sequences used for the real time quantitative PCR/RT-qPCR
References
3. B├©gue-Kirn C, Smith AJ, Lesot H, et al. : Comparative analysis of TGF beta s, BMPs, IGF1, msxs, fibronectin, osteonectin and bone sialoprotein gene expression during normal and in vitro-induced odontoblast differentiation. Int J Dev Biol, 38:405-420, 1994.

4. Papagerakis P, Berdal A, Macdougall M, et al. : Investigation of osteocalcin, osteonectin, and dentin sialophosphoprotein in developing human teeth. Bone, 30:377-385, 2002.


5. Gronthos S, Mankani M, Shi S, et al. : Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA, 97:13625-13630, 2000.



6. Lindroos B, M├żenp├ż├ż K, Miettinen S, et al. : Characterisation of human dental stem cells and buccal mucosa fibroblasts. Biochem Biophys Res Commun, 368:329-335, 2008.


7. Miura M, Gronthos S, Shi S, et al. : SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA, 100:5807-5812, 2003.



8. Huang AH, Chen YK, Chan AW, et al. : Isolation and characterization of dental pulp stem cells from a supernumerary tooth. J Oral Pathol Med, 37:571-574, 2008.


9. Sun HJ, Bahk YY, Lee JW, et al. : A proteomic analysis during serial subculture and osteogenic differentiation of human mesenchymal stem cell. J Orthop Res, 24:2059-2071, 2006.


10. Kim JS : Characterization of Differentiation of the Supernumerary Dental Pulp Stem Cells toward the Odontoblast by Application Period of Additives. J Korean Acad Pediatr Dent, 42:312-318, 2015.

11. Primosch RE : Anterior supernumerary teeth-assessment and surgical intervention in children. Pediatr Dent, 3:204-215, 1981.

12. Huang WH, Tsai TP, Su HL : Mesiodens in the primary dentition stage: a radiographic study. ASDC J Dent Child, 59:186-189, 1992.

13. Kim JB : Managing Complications Related to Multiple Supernumerary Teeth. J Korean Acad Pediatr Dent, 41:180-186, 2014.

14. Gronthos S, Brahim J, Shi S, et al. : Stem cell properties of human dental pulp stem cells. J Dent Res, 81:531-535, 2002.


15. Bruder SP, Jaiswal N, Haynesworth SE : Growth kinetics, self-renewal, and the osteogenic potential of purified human mesenchymal stem cells during extensive subcultivation and following cryopreservation. J Cell Biochem, 64:278-294, 1997.


16. Digirolamo CM, Stokes D, Prockop DJ, et al. : Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol, 107:275-281, 1999.


17. Min JH, Ko SY, Jang YJ, et al. : Dentinogenic potential of human adult dental pulp cells during the extended primary culture. Hum Cell, 24:43-50, 2011.


18. Livak KJ, Schmittgen TD : Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25:402-408, 2001.


19. Seo BM, Miura M, Shi S, et al. : Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet, 364:149-155, 2004.


20. Morsczeck C, G├Čtz W, Hoffmann KH, et al. : Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol, 24:155-165, 2005.


21. Sonoyama W, Liu Y, Shi S, et al. : Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS One, 1:79, 2006.

22. Huang W, Tsai T, Su H : Mesiodens in the primary dentition stage: a radiographic study. ASDC J Dent Child, 59:186-189, 1991.
23. Kamata N, Fujimoto R, Yasumoto S, et al. : Immortalization of human dental papilla, dental pulp, periodontal ligament cells and gingival fibroblasts by telomerase reverse transcriptase. J Oral Pathol Med, 33:417-423, 2004.


24. Conget PA, Minguell JJ : Phenotypical and functional properties of human bone marrow mesenchymal progenitor cells. J Cell Physiol, 181:67-73, 1999.


25. Aubin JE, Liu F, Gupta AK, et al. : Osteoblast and chondroblast differentiation. Bone. 17:S77-S83, 1995.

26. Arana-Chavez VE, Massa LF : Odontoblasts: the cells forming and maintaining dentine. Int J Biochem Cell Biol, 36:1367-1373, 2004.


27. Termine JD, Kleinman HK, Martin GR, et al. : Osteonectin, a bone-specific protein linking mineral to collagen. Cell, 26:99-105, 1981.


28. Wasi S, Otsuka K, Termine JD, et al. : An osteonectinlike protein in porcine periodontal ligament and its synthesis by periodontal ligament fibroblasts. Can J Biochem Cell Biol, 62:470-478, 1984.


29. Garcia JM, Martins MD, Marques MM, et al. : Immunolocalization of bone extracellular matrix proteins (type I collagen, osteonectin and bone sialoprotein) in human dental pulp and cultured pulp cells. Int Endod J, 36:404-410, 2003.


30. Lauc G, Heffer-Lauc M : Shedding and uptake of gangliosides and glycosylphosphatidylinositol-anchored proteins. Biochim Biophys Acta, 1760:584-602, 2006.


31. Hanawa M, Takano Y, Wakita M : An autoradiographic study of calcium movement in the enamel organ of rat molar tooth germs. Arch Oral Biol, 35:899-906, 1990.


32. Bronckers AL, Price PA, Karsenty G, et al. : Studies of osteocalcin function in dentin formation in rodent teeth. Eur J Oral Sci, 106:795-807, 1998.


33. Karsenty G : Role of Cbfa1 in osteoblast differentiation and function. Semin Cell Dev Biol, 11:343-346, 2000.


34. George A, Sabsay B, Veis A, et al. : Characterization of a novel dentin matrix acidic phosphoprotein. J Biol Chem, 268:12624-12630, 1993.

35. D'souza RN, Cavender A, MacDougall M, et al. : Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res, 12:2040-2049, 1997.


- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 862 View
- 73 Download
- Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print